Development of High-Throughput DNA Sequencing Techniques to Improve and Advance Environmental Monitoring and Bioassessment
 Scientists can learn about the health of rivers, streams, lakes, and other aquatic ecosystems by looking at the species that live in them – the populations of insects, snails, and worms that can be found in different aquatic ecosystems can indicate the overall health of the areas they live in. However, finding and evaluating these species can be difficult, costly, and time-consuming, resulting in delays in decisions related to the protection of these ecosystems.
Scientists can learn about the health of rivers, streams, lakes, and other aquatic ecosystems by looking at the species that live in them – the populations of insects, snails, and worms that can be found in different aquatic ecosystems can indicate the overall health of the areas they live in. However, finding and evaluating these species can be difficult, costly, and time-consuming, resulting in delays in decisions related to the protection of these ecosystems.
Currently, biological monitoring programs for rivers, streams and other aquatic areas focus primarily on identifying aquatic macroinvertebrates such as insects, snails and worms. These efforts can take 6 to 12 months. However, because these aquatic organisms leave molecular-level DNA "footprints" in the environments where they live, scientists can use molecular methods such as DNA barcoding to obtain reliable information about the species in a given area. DNA barcoding uses short genetic markers in an organism's DNA to identify it as belonging to a particular species. It is also be possible to use these DNA-based techniques to accurately identify much smaller aquatic organisms, such as algae and bacteria.
EPA scientists are now using new molecular technology, such as high-throughput DNA sequencing to monitor and assess aquatic ecosystems. Sequencing technology is used to determine the exact order of nucleotides in a DNA sample. Nucleotides are the building blocks of DNA. With this cutting-edge technology, scientists can process multiple water samples simultaneously and create a comprehensive list of organisms present at that site. In fact, samples from up to approximately 400 sites could be analyzed simultaneously, which would drastically reduce laboratory testing costs.
EPA scientists are developing a method to provide robust, reliable and timely bioassessments for samples from streams and marine coastal sites. Using DNA sequencing technology, they are working on ways to identify large invertebrate species and comparing their results with the findings of taxonomists who have identified the species individually. For the coastal samples, they are looking at soil samples to see if they can identify large and small invertebrate species using DNA barcoding techniques.
Because these methods will provide more data than standard bioassessment protocols, EPA scientists think these new techniques will not only help determine the condition of specific aquatic ecosystems, but might also detect whether a site is improving or degrading. These methods may also help scientists detect the presence of pollutants in streams.
In the figure below, the likely benefits of using genomic community data are blue. The darkly-shaded boxes indicate the areas where current efforts are focused.
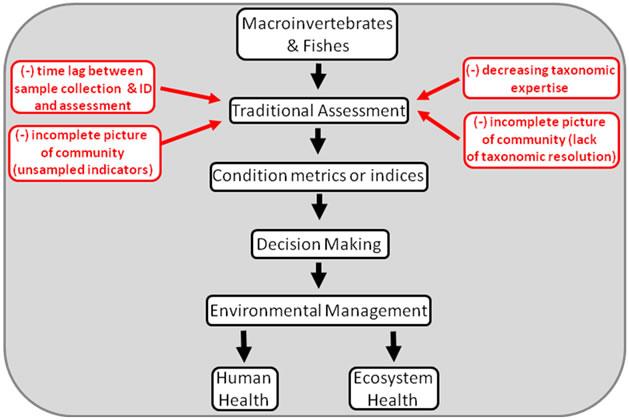 Figure 1. Conceptual model of traditional bioassessment programs. Problems related to traditional morphological taxonomic identification are marked in red.
Figure 1. Conceptual model of traditional bioassessment programs. Problems related to traditional morphological taxonomic identification are marked in red.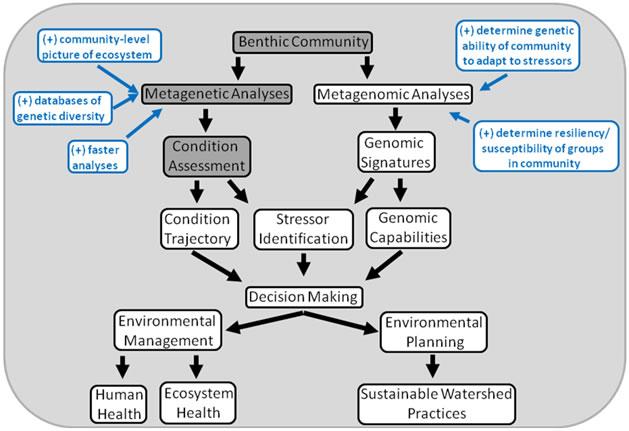 Figure 2. Conceptual model of the future of bioassessment and other community-level environmental analyses. Much of this work is still hypothetical, although the technology exists to begin developing this framework. Likely benefits of using genomic community data are in blue. The darkly shaded boxes denote the areas where our current efforts are focused.
Figure 2. Conceptual model of the future of bioassessment and other community-level environmental analyses. Much of this work is still hypothetical, although the technology exists to begin developing this framework. Likely benefits of using genomic community data are in blue. The darkly shaded boxes denote the areas where our current efforts are focused.
Publications:
- Does DNA barcoding improve performance of traditional stream bioassessment metrics? Eric.D. Stein, Bryan P. White, Raphael D. Mazor, John K. Jackson, Juliann M. Battle, Peter E. Miller, Erik M. Pilgrim, and Bernard W. Sweeney. Freshwater Science. 2014, 33(1): 302-311.
- Cryptic biodiversity in streams: a comparison of macroinvertebrate communities based on morphological and DNA barcode identifications. John K. Jackson, Juliann M. Battle, Bryan P. White, Erik M. Pilgrim, Eric D. Stein, Peter E. Miller, and Bernard W. Sweeney. Freshwater Science. 2014, 33(1): 312-324.
- Incorporation of DNA barcoding into a large-scale biomonitoring program: opportunities and pitfalls. Erik M. Pilgrim, Suzanne A. Jackson, Stephanie Swenson, Istvan Turcsanyi, Ellen Friedman, Lee Weigt, and Mark J. Bagley. J. N. Am. Benthol. Soc. 2011, 30(1): 217-231.
- Can DNA barcodes of stream macroinvertebrates improve descriptions of community structure and water quality? Bernard W. Sweeney, Juliann M. Battle, and John K. Jackson. J. N. Am. Benthol. Soc. 2011, 30(1): 195-216.
- Environmental Barcoding: A Next-Generation Sequencing Approach for Biomonitoring Applications Using River Benthos. Mehrdad Hajibabaei, Shadi Shokralla, Xin Zhou, Gregory A. C. Singer, Donald J. Baird. PLoS ONE. 2011, 6(4): e17497, 1-7.
Top Questions:
- How does DNA barcoding work? Exit
- How do we use aquatic organisms to measure ecosystem health?
- Ask a Question
Related Topics:
- EPA, Notre Dame researchers discuss challenges in adopting DNA-based methods for monitoring invasive species in U.S. water bodies
- Conceptual diagrams of bioassessment research
Featured Resource:
- National Rivers and Streams Assessment Fact Sheet (2pp, 0.1 MB, About PDF)
